Human beings have twenty-three pairs of chromosomes in each of their body cells. Most bacteria are much smaller and simpler than human cells, and generally have only one chromosome. In addition to this single chromosome, bacteria often carry plasmids. Plasmids like chromosomes are made of genetic material but, unlike chromosomes, are non-essential. However, a plasmid, which is small ring of DNA, can change the characteristics of bacteria. It can make a bacterium resistant to antibiotics, for instance. This is important if the bacterium happens to cause a disease and a doctor wants to cure the sick patient with antibiotics.
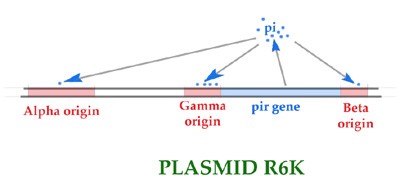
While obtaining his Ph.D. in Bacteriology from the University of Wisconsin-Madison in 1995, Frank Wu studied the antibiotic-resistance plasmid R6K in the bacterium E. coli., which is commonly found in the human intestines. Wu particularly studied the control of replication of R6K. R6K replication occurs at three sites on the plasmid called the alpha, gamma and beta origins. Replication also requires a protein, called pi, which is produced from (encoded by) R6K itself. Pi protein binds to sites in all three origins.

The gamma origin consists of several adjacent segments, including a region bound by pi protein. The origin also contains sites bound by additional proteins which are produced by the host bacterial cell, but which the plasmid co-ops in its own reproduction. Some of these host proteins are DnaA, IHF and Fis. DnaA binds to sites known as "boxes" and is involved in DNA replication. IHF and Fis also bind to specific sites; they bend DNA and are involved in DNA replication and expression of various genes. pi protein can be either "wild-type" (e.g., "normal") or "copy-up." The latter cause an increase in replication of plasmid R6K. R6K plasmids are normally present in a cell at 15-30 copies per cell, but "copy-up" pi mutants allow a huge increase in the copy number (up to 10-fold). Pi can also be produced in the cell at a low or high level; low levels of pi also increase replication (e.g., the number of plasmids per cell) relative to the high level of pi.
1. Roles of a 106-bp origin enhancer and Escherichia coli DnaA protein in replication of plasmid R6K
Frank Wu, Ilya Goldberg, and Marcin Filutowicz, Nucleic Acids Research, 20:811-817 (1992)
A dnaA "null" strain could not support replication of intact plasmid R6K or derivatives containing combinations of its three replication origins (alpha, gamma, beta). DnaA binds in vitro to sites in two functionally distinct segments of the central gamma origin. The 277-bp core segment is common to all three origins and contains DnaA box 2, which cannot be deleted without preventing replication. Immediately to the left of the core lies the 106-bp origin enhancer, which contains DnaA box 1. When the origin enhancer is deleted, the core alone can still initiate replication if levels of wild-type pi protein are decreased or if copy-up pi mutant proteins are provided in trans. DnaA does not effect expression of R6K replication initiator protein pi, although several DnaA boxes were identified in the coding segment of the pir gene, which encodes pi. Together these data suggest that a single DnaA box, 2, is sufficient for initiation from the gamma origin and might be sufficient and required for the activity of the alpha and beta origins as well. Implications of the DnaA protein binding to two domains of the gamma origin and the role of the 106-bp origin enhancer in replication are discussed.
2. Binding of DnaA protein to a replication enhancer counteracts the inhibition of plasmid R6K gamma origin replication mediated by elevated levels of R6K pi protein
Frank Wu, Igor Levchenko, and Marcin Filutowicz, Journal of Bacteriology, 176:6795-6801 (1994)
Replication of the gamma origin of Escherichi coli plasmid R6K requires pi protein, encoded by the R6K pir gene and many host factors, including DnaA protein. Pi has dual roles, activating replication at low levels and inhibiting replication at high levels. The inhibitory function of pi is counteracted by integration host factor (IHF) and a specific sequence of the origin called the enhancer. This 106-bp DNA segment contains a binding site for DnaA protein (DnaA box 1). In this study, we mutated this site to determine if it was required for the enhancer's function. Using gamma origin derivative plasmids with the DnaA box 1 altered or deleted, we show that this site is necessary to protect the origin against levels of wild-type pi protein that would otherwise inhibit replication. To show that the base substitutions in DnaA box 1 weakened the binding of DnaA, we developed a new application of the agarose gel retardation assay. This quick and easy assay has broad applicability, as shown in binding studies with DNA fragments carrying a different segment of the R6K origin, the chromosomal origin (oriC), or the pUC origin. The gel retardation assay suggests a stoichiometry of DnaA binding different from that deduced from other assays.
3. A DNA segment conferring stable maintenance on R6K gamma-origin core replicons
Frank Wu, Igor Levchenko, and Marcin Filutowicz, Journal of Bacteriology, 177: 6338-6345 (1995)
The plasmid R6K gamma origin consists of two adjacent modules, the enhancer and the core, and requires R6K initiator protein pi for replication. While the core alone can replicate at a low level of wild-type pi protein, we show here that host cells do not stably maintain core plasmids. The presence of the enhancer segment confers stable inheritance on core plasmids without a significant change in average plasmid copy number. Deletions and site-directed mutatgenesis indicated that the stability of core plasmids is not mediated by binding sites or consensus sequences in the enhancer for DnaA, pi protein, gyrase, Fis, or Dcm methylase. Proper segregation of core plasmids requires only the R6K stb or stability-related region, which includes the 20-bp segment of the 100-bp enahncer adjacent to the core. The use of the pi116 copy-up pi mutant protein, which increases plasmid copy number four-fold, does not stabilize core plasmids lacking the enhancer. We also show that at an elevated level of wild-type pi protein, the gamma origin plasmid is unstable, even in the presence of the enhancer. We discuss the differences and similarities between the R6K stability system and those found in other plasmids.
4. Preponderance of Fis-binding sites in the R6K gamma origin and the curious effect of the penicillin resistance marker on replication of this origin in the absence of Fis
Frank Wu, Jiazhen Wu, Jennifer Ehley, and Marcin Filutowicz, Journal of Bacterioogy, 178: 4965-4974 (1996)
Fis protein is shown here to bind to 10 sites in the gamma origin of plasmid R6K. This Fis-binding sites overlap all the previously identified binding sites in the gamma origin for the plasmid-encoded pi initiator protein and three host-encoded proteins, DnaA, integration host factor (IHF), and RNA polymerase. However, the requirement of Fis for R6K replication depends on the use of copy-up pi-protein variants and, oddly, the antibiotic resistance marker on the plasmid. In Fis-deficient cells, copy-up pi variants cannot drive replication of R6K gamma-origin plasmids carrying the bla gene encoding resistance to penicillin (penR), but can drive replication of plasmids with the same origin but carrying the chloramphenicol acetyltransferase gene encoding chroamphenicol resistance (CmR). In contrast, R6K replication driven by wild-type pi is unaffected by the antibiotic resistance marker in the absence of Fis protein. Individually, none of these elements (copy-up pi, Fis deficiency, or drug markers) prevents R6K replication. The replication defect is not caused by penicillin in the medium or runaway replication and is unaffected by the orientation of the bla gene relative to the origin. Replication remains inhibited when part of the bla coding segment is deleted but the bla promoter is left intact. However, replication is restored by insertion of transcriptional terminators on either side of the gamma origin, suggesting that excess transcription from the bla gene may inactive replication driven by pi copy-up mutants in the absence of Fis. This study suggests that vector sequences such as drug markers may not be inconsequential in replication studies, as is generally assumed.
Back to Science Fiction and Humor Writing
Back to The Official Frank Wu Website homepage